Table of Contents
Muscles Overview
Visual3D allows users to define muscles as linear segments connecting a series of landmarks. These landmarks can be created using any of the rules for creating landmarks in Visual3D.
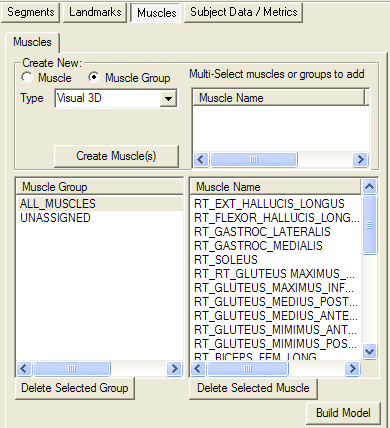
Muscles are created and edited from the Muscle Tab in Model Builder Mode.
Muscle Definition
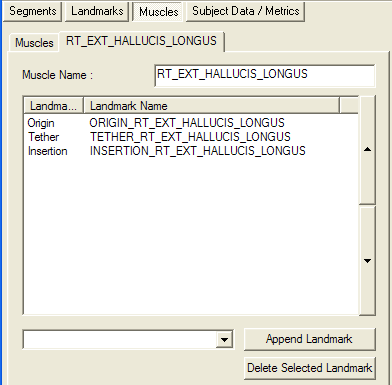
Muscles are defined as a connect series of linear segments, passing from Landmark to Landmark. The user can add or delete the landmarks in the list box. Note that the order of the landmarks is important regarding how they are interpreted:
- the first landmark is always considered to be the Origin of the muscle;
- the last landmark is always considered to be the Insertion of the muscle; and
- all other landmarks are considered to be Tether (Via) points and are connected in the order they appear in the list box.
Terry Database
The Terry Database comes from an article by Kepple et al. in 1998. The original dataset can be downloaded here.
The landmarks are stored in ASCII files. The structure of the dataset is described in the files. An excerpt from the femur landmarks is presented here:
- Description of landmarks digitized on the femur
- Each landmark is listed in the same order as they appear in the corresponding model files. The model file format is:
- (landmark #) x y z (# of specimens used to generate norm landmark)
- x is approximately anterior, y is superior, and z is right
- units are mm
This file contains for each landmark:
- landmark number followed by a description of where the landmark is located.
- landmark type. The landmark type can be either externally palpable, muscle origin, muscle insertion, muscle connector (to wrap muscle around bone or through retinaculum), joint center (assumed to be a point of contact with the adjacent segment), or anthropometric (a point which may be of value for traditional anthropology measurements).
- list of muscles associated with the point (origin, insertion or connector).
Affine Scaling of a Terry Landmark
The linear algebra technique described in Sommer, Miller, and Pihanowske (1982), termed affine scaling, scales the data based on all measured landmarks and allows for the statistical accumulation of landmark data in order to generate normative models.
Each landmark is defined relative to 3 or 4 landmarks that were selected to be palpable on living subjects. For example, the thigh landmarks are defined relative to the greater trochanter, the lateral epicondyle, and the medial epicondyle.
Visual3D Representation of a Terry Landmark
The following Visual3D command is used to define a landmark representing the Origin of the Right Medial Gastrocnemius Muscle based on Affine Scaling
Add_Landmark_Using_Affine_Scaling /VISUAL3D_LANDMARK_NAME=ORIGIN_RT_GASTROC_MED /CALIBRATION_FILE=*model.mdh /LINKMODEL_MARKER_NAMES=RGT+RLK+RMK /LANDMARK_LOCATION_IN_LOCAL_COORDINATE_SYSTEM=-26.117+-381.432+-24.205 /SEGMENT_NAME=RTH /MARKER1_IN_LOCAL_COORDINATE_SYSTEM=0+0+0 /MARKER2_IN_LOCAL_COORDINATE_SYSTEM=0+-380.878+43.317 /MARKER3_IN_LOCAL_COORDINATE_SYSTEM=0+-394.841+-43.317 ! /MARKER4_IN_LOCAL_COORDINATE_SYSTEM= /SCALE_ML=FALSE /SCALE_AXIAL=TRUE ;
Note that the motion capture cap markers and/or landmarks for the three homologous anatomical points must exist in the model file.
LGT= Left Greater Trochanter LLK= Left Lateral Knee (epicondyle) LMK= Left Lateral Knee (epicondyle)
In the above example, the Terry information for the thigh segments (femur.txt) required are the following:
- most lateral point on greater trochanter
- type:palpable landmark
- muscle:
- most lateral point on lateral epicondyle
- type:palpable landmark
- muscle:
- most medial point on medial epicondyle
- type:palpable landmark
- muscle:
- most superior point on medial condyle
- type: muscle origin
- muscle: origin of Gastrocnemius medial head
The numerical values associated with these locations for a White Male are stored in the file femur.mod
FEMUR - right and left - 26 males, 26 females 41 LANDMARKS
| 1 | 0.000 | 0.000 | 0.000 | 104.000 |
| 2 | 0.000 | -380.878 | 43.317 | 104.000 |
| 3 | 0.000 | -394.841 | -43.317 | 104.000 |
| ….. | ||||
| 30 | -26.117 | -381.432 | -24.205 | 104.000 |
References
1. Kepple TM, Sommer HJ, Siegel KL, Standhope SJ (1998) A three-dimensional musculoskeletal database for the lower extremities. Journal of Biomechanics 31, pp 77-80 DOI
The locations of idealized muscle attachments on the pelvis, both femurs, both tibias and fibulas, and both feet were accurately digitized for 52 dried skeletal specimens….Statistical accumulation and scaling techniques were used to generate highly representative normative models, which were divided into groups and tested for differences based on gender and race….Containing over 12000 anatomical landmarks digitized from 52 dried skeletons, this study represents an improvement over previous databases by an order of magnitude
Note that the limitation of the data set is that because the data collection was from bones, there is little information on muscle path. For example, it was not possible to identify tether (or via) points related to, for example, attachments to the patella. Careful estimation of these via points is possible, but it is difficult to verify the accuracy of these estimates. This challenge is most serious for the estimate of the moment arms around some joints, and for the estimate of length of muscles that are short relative to the actual path.
If the purpose the muscle representation is visualization (e.g. mapping EMG to the color of the rendered muscles) these issues are not relevant.
2. Sommer H, Miller N, Pihanowske G (1982) Three-dimensional osteometric scaling and normative modelling of skeletal segments. Journal of Biomechanics 15, pp 171-180. DOI
Many analytical biomechanics methods require extensive three-dimensional descriptions of anatomical geometry. In particular, researchers requiring the three-dimensional coordinates of specific boney landmarks (e.g. tendon and ligament attachments) are often forced to extrapolate such measurements from an experimental specimen set to their subject geometry.
This work offers an approach to two problems inherent above; accurate extrapolation of specimen landmark locations to subject homologues and statistical accumulation of normative three-dimensional anatomical landmark data bases. A least squares solution for an affine scaling transformation from specimen to subject is used which incorporates both right-left and same hand comparisons. A two stage technique is formulated to consecutively remove landmark location variation and to size a normative specimen from a set of similar specimens. This ability to statistically represent a specimen set will provide better geometric models for other analytical studies and prosthetic design and evaluation.
Example Data Set with Meta Commands and Pipeline
This is example demonstrates how to use a dataset compatible with the Terry Database within Visual3D.
Prepare
- Download the example dataset.
- Download the Meta-Commands and place them in a folder labelled Meta-Commands inside the Visual3D plugins folder
- Download the example Visual3D pipeline. Note to download this file, right mouse click on the link and save the link to your computer.
Steps
- Load the cmo file into Visual3D
- Execute the pipeline to create Visual3D representations of a collection of the Terry Database muscle.
Note that the first command of this example pipeline specifies the FOLDER containing the data. You will need to recreate this folder path or specify the path on your computer.
Visual3D Meta-Command for a Terry Muscle
As an example, the Gastrocnemius muscle can be defined using the following Meta-Command
! BEGIN_META ! META_CMD_NAME=Terry_Rt_Plantar_Flexors_Group ! META_PARAM= MODEL_NAME : string ::yes ! META_PARAM= RIGHT_THIGH_MARKERS : string ::yes ! META_PARAM= RIGHT_SHANK_MARKERS: string ::yes ! META_PARAM= RIGHT_FOOT_MARKERS: string ::yes ! END_META ! ------------------------------------------------------------- ! Gastrocnemius\\ ! ------------------------------------------------------------- Add_Landmark_Using_Affine_Scaling /VISUAL3D_LANDMARK_NAME=ORIGIN_RT_GASTROC_MED /CALIBRATION_FILE=::MODEL_NAME /LINKMODEL_MARKER_NAMES=::RIGHT_THIGH_MARKERS /LANDMARK_LOCATION_IN_LOCAL_COORDINATE_SYSTEM=-26.117+-381.432+-24.205 /SEGMENT_NAME=RTH /MARKER1_IN_LOCAL_COORDINATE_SYSTEM=0+0+0 /MARKER2_IN_LOCAL_COORDINATE_SYSTEM=0+-380.878+43.317 /MARKER3_IN_LOCAL_COORDINATE_SYSTEM=0+-394.841+-43.317 ! /MARKER4_IN_LOCAL_COORDINATE_SYSTEM= /SCALE_ML=FALSE /SCALE_AXIAL=TRUE ; Add_Landmark_Using_Affine_Scaling /VISUAL3D_LANDMARK_NAME=TETHER_RT_GASTROC_MED /CALIBRATION_FILE=::MODEL_NAME /LINKMODEL_MARKER_NAMES=::RIGHT_THIGH_MARKERS /LANDMARK_LOCATION_IN_LOCAL_COORDINATE_SYSTEM=-37.929+-401.247+-20.216 /SEGMENT_NAME=RTH /MARKER1_IN_LOCAL_COORDINATE_SYSTEM=0+0+0 /MARKER2_IN_LOCAL_COORDINATE_SYSTEM=0+-380.878+43.317 /MARKER3_IN_LOCAL_COORDINATE_SYSTEM=0+-394.841+-43.317 ! /MARKER4_IN_LOCAL_COORDINATE_SYSTEM= /SCALE_ML=FALSE /SCALE_AXIAL=TRUE ; Add_Landmark_Using_Affine_Scaling /VISUAL3D_LANDMARK_NAME=INSERTION_RT_GASTROC_MED /CALIBRATION_FILE=::MODEL_NAME /LINKMODEL_MARKER_NAMES=::RIGHT_FOOT_MARKERS /LANDMARK_LOCATION_IN_LOCAL_COORDINATE_SYSTEM=-0.529+1.120+-0.726 /SEGMENT_NAME=RFT /MARKER1_IN_LOCAL_COORDINATE_SYSTEM=0+0+0 /MARKER2_IN_LOCAL_COORDINATE_SYSTEM=85.524+0+-35.699 /MARKER3_IN_LOCAL_COORDINATE_SYSTEM=88.645+0+35.699 ! /MARKER4_IN_LOCAL_COORDINATE_SYSTEM= /SCALE_ML=FALSE /SCALE_AXIAL=FALSE ; Add_Landmark_Based_Muscle /MUSCLE_NAME=RT_GASTROC_MEDIALIS /LANDMARK_ORIGIN=ORIGIN_RT_GASTROC_MED /LANDMARK_TETHERS=TETHER_RT_GASTROC_MED /LANDMARK_INSERTION=INSERTION_RT_GASTROC_MED /LINK_MODEL_NAME=::MODEL_NAME ; Add_Landmark_Using_Affine_Scaling /VISUAL3D_LANDMARK_NAME=ORIGIN_RT_GASTROC_LAT_PLANTARIS /CALIBRATION_FILE=::MODEL_NAME /LINKMODEL_MARKER_NAMES=::RIGHT_THIGH_MARKERS /LANDMARK_LOCATION_IN_LOCAL_COORDINATE_SYSTEM=-20.936+-373.101+18.664 /SEGMENT_NAME=RTH /MARKER1_IN_LOCAL_COORDINATE_SYSTEM=0+0+0 /MARKER2_IN_LOCAL_COORDINATE_SYSTEM=0+-380.878+43.317 /MARKER3_IN_LOCAL_COORDINATE_SYSTEM=0+-394.841+-43.317 ! /MARKER4_IN_LOCAL_COORDINATE_SYSTEM= /SCALE_ML=FALSE /SCALE_AXIAL=TRUE ; Add_Landmark_Using_Affine_Scaling /VISUAL3D_LANDMARK_NAME=TETHER_RT_GASTROC_LAT_PLANTARIS /CALIBRATION_FILE=::MODEL_NAME /LINKMODEL_MARKER_NAMES=::RIGHT_THIGH_MARKERS /LANDMARK_LOCATION_IN_LOCAL_COORDINATE_SYSTEM=-30.662+-390.245+23.209 /SEGMENT_NAME=RTH /MARKER1_IN_LOCAL_COORDINATE_SYSTEM=0+0+0 /MARKER2_IN_LOCAL_COORDINATE_SYSTEM=0+-380.878+43.317 /MARKER3_IN_LOCAL_COORDINATE_SYSTEM=0+-394.841+-43.317 ! /MARKER4_IN_LOCAL_COORDINATE_SYSTEM= /SCALE_ML=FALSE /SCALE_AXIAL=TRUE ; Add_Landmark_Using_Affine_Scaling /VISUAL3D_LANDMARK_NAME=INSERTION_RT_GASTROC_LAT_PLANTARIS /CALIBRATION_FILE=::MODEL_NAME /LINKMODEL_MARKER_NAMES=::RIGHT_FOOT_MARKERS /LANDMARK_LOCATION_IN_LOCAL_COORDINATE_SYSTEM=-0.529+1.120+-0.726 /SEGMENT_NAME=RFT /MARKER1_IN_LOCAL_COORDINATE_SYSTEM=0+0+0 /MARKER2_IN_LOCAL_COORDINATE_SYSTEM=85.524+0+-35.699 /MARKER3_IN_LOCAL_COORDINATE_SYSTEM=88.645+0+35.699 ! /MARKER4_IN_LOCAL_COORDINATE_SYSTEM= /SCALE_ML=FALSE /SCALE_AXIAL=FALSE ; Add_Landmark_Based_Muscle /MUSCLE_NAME=RT_GASTROC_LATERALIS /LANDMARK_ORIGIN=ORIGIN_RT_GASTROC_LAT_PLANTARIS /LANDMARK_TETHERS=TETHER_RT_GASTROC_LAT_PLANTARIS /LANDMARK_INSERTION=INSERTION_RT_GASTROC_LAT_PLANTARIS /LINK_MODEL_NAME=::MODEL_NAME\\ ;
Pipeline Command for Above Meta-Command
A meta-command is a Visual3D pipeline into which parameters can be passed. The meta-command should be stored in a folder labeled Meta-Commands, which is located in the Visual3D Plugins Folder.
The header in the meta-command defines the data that should be passed:
! BEGIN_META ! META_CMD_NAME=Terry_Rt_Plantar_Flexors_Group ! META_PARAM= MODEL_NAME : string ::yes ! META_PARAM= RIGHT_THIGH_MARKERS : string ::yes ! META_PARAM= RIGHT_SHANK_MARKERS: string ::yes ! META_PARAM= RIGHT_FOOT_MARKERS: string ::yes ! END_META
This command will appear in the Visual3D Pipeline Dialog list box in a folder labeled Meta_Commands. The command will be similar to the following:
Terry_Rt_Plantar_Flexors_Group /MODEL_NAME= *model.mdh /RIGHT_THIGH_MARKERS= RGT+RLK+RMK /RIGHT_SHANK_MARKERS= LTT +RLA+RMA /RIGHT_FOOT_MARKERS= RHL+RFT2+RFT1 ;